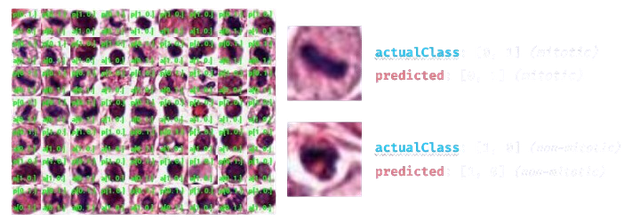
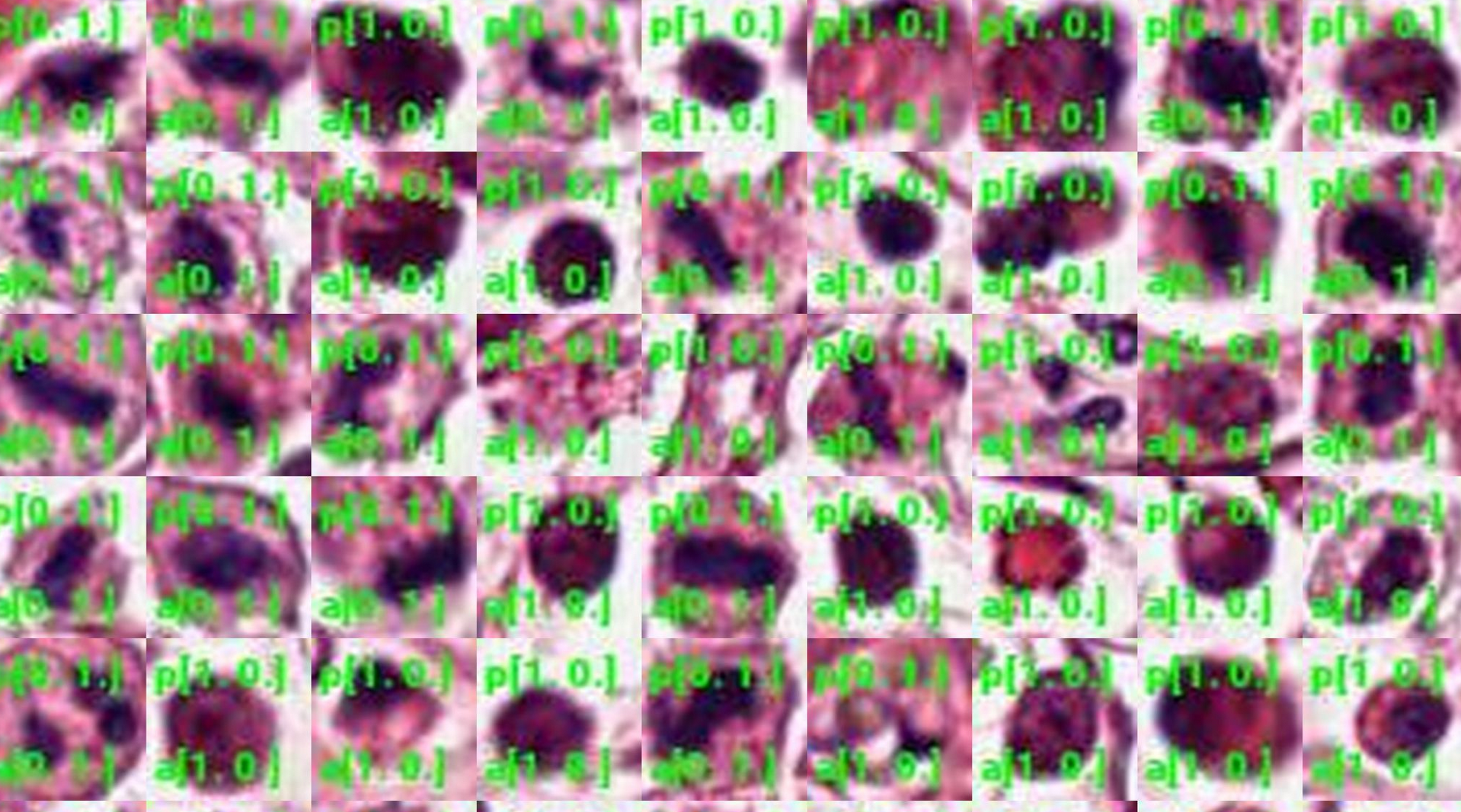
Goal
Given an image of a cell, determine whether or not it’s going through mitosis.
Data processing
All data came from this Kaggle dataset. The dataset provides DICOM files, which in this case, are massive resolution images of tissue samples where individual cells can be identified. In a separate SQL database, there are the coordinates and annotations of mitotic state for all of the cells.
After obtaining all cells that match the agreedClass, slide, limit, and size, I then split the data into train/test sets.
The Keras model built for this was a pretty simple CNN, but it got the job done.
(train_images, train_labels), (test_images, test_labels), (limit, train_percent) = generate_final_data()
model = keras.Sequential()
model.add(layers.Conv2D(32, (3, 3), activation="relu", input_shape=(40, 40, 3)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation="relu"))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation="relu"))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation="relu"))
model.add(layers.Flatten())
model.add(layers.Dense(64, activation="relu"))
model.add(layers.Dense(2, activation="softmax"))Results
After running the model for epochs=7 and batch_size=30, I obtained ~94% accuracy! The final results were:
=================================================================
Total params: 129,570
Trainable params: 129,570
Non-trainable params: 0
_________________________________________________________________
188/188 [==============================] - 1s 4ms/step - loss: 0.1297 - accuracy: 0.9467
0.9466666579246521 <~ accuracy
0.12974701821804047 <~ loss